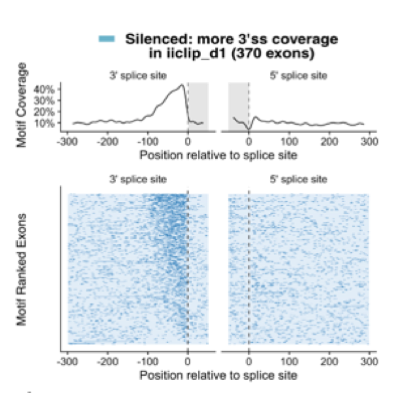
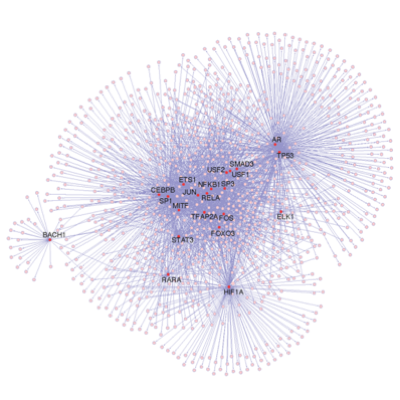
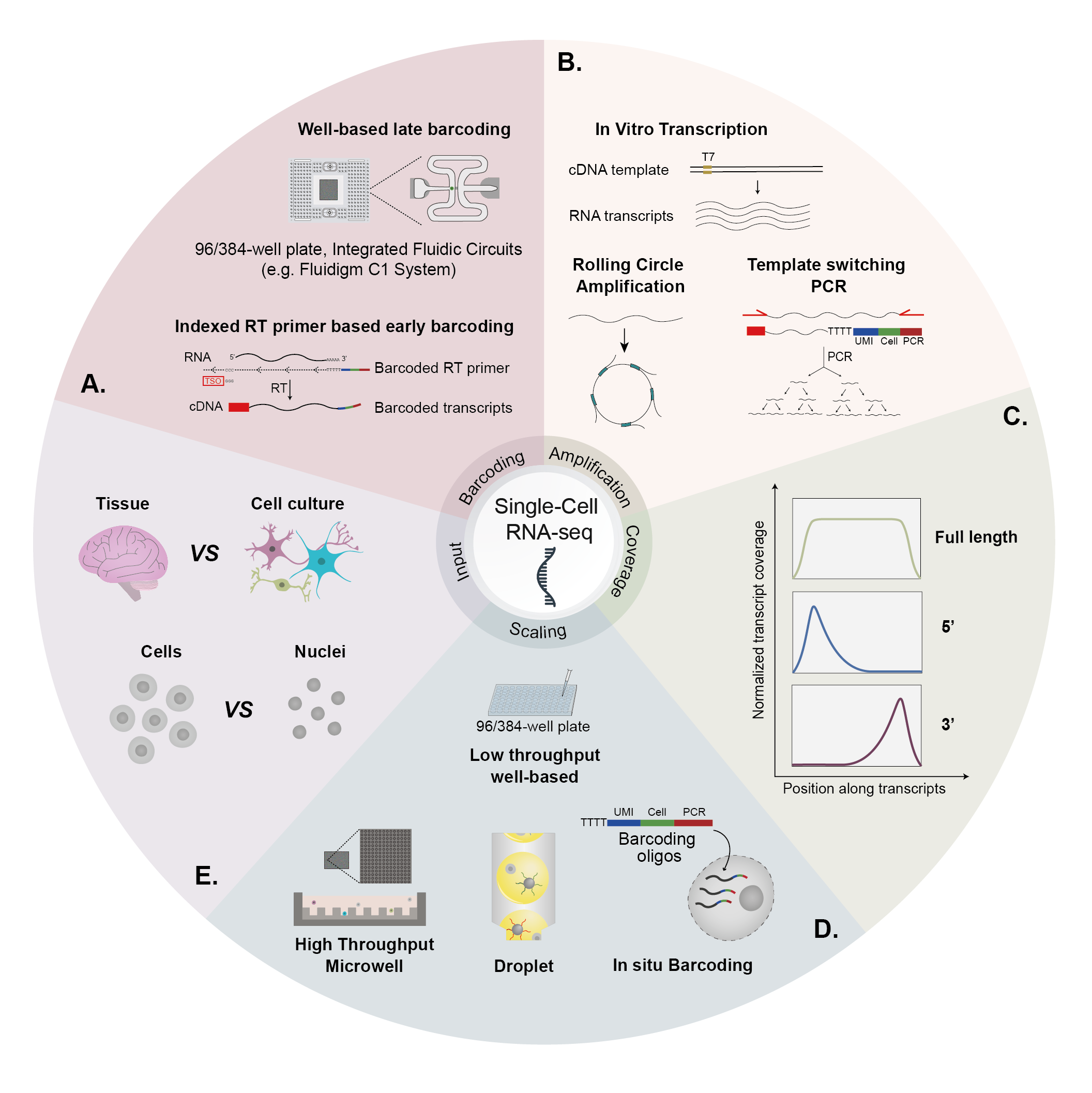
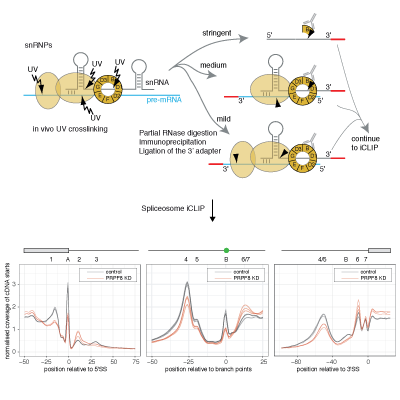
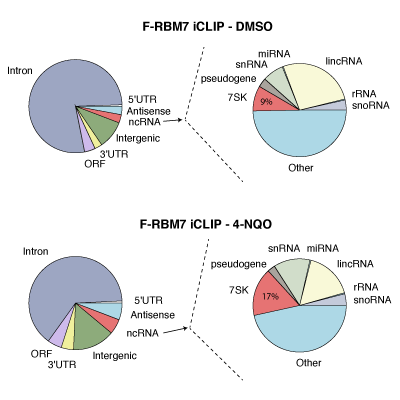
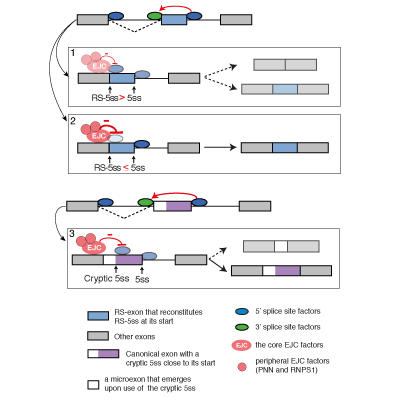
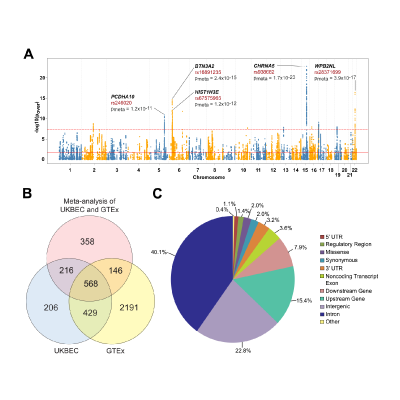
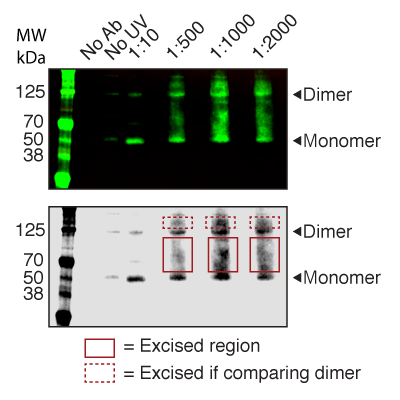
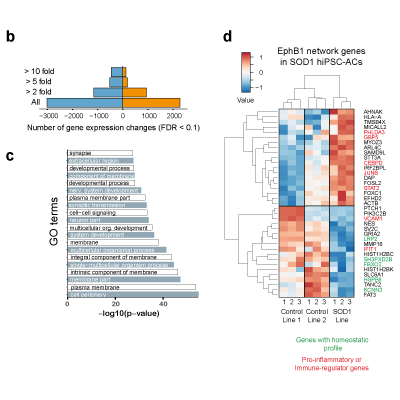
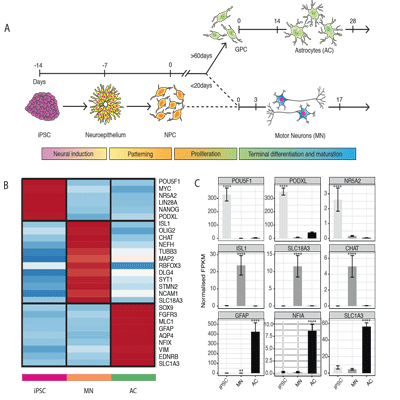
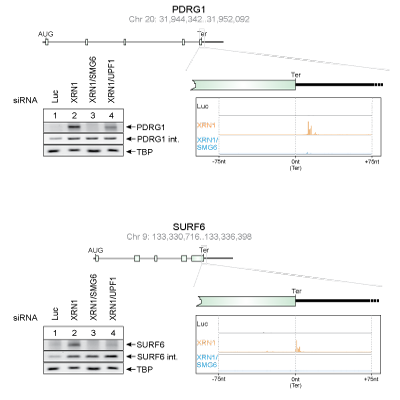
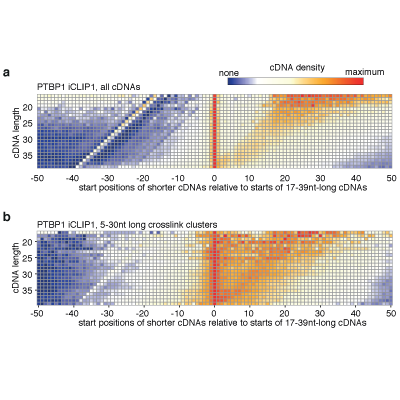
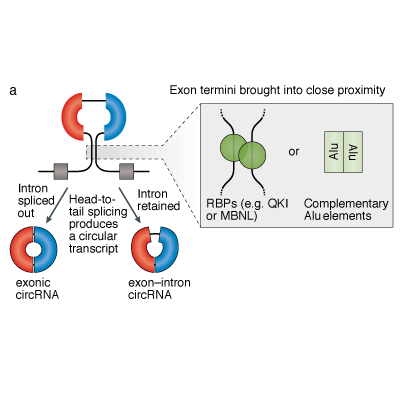
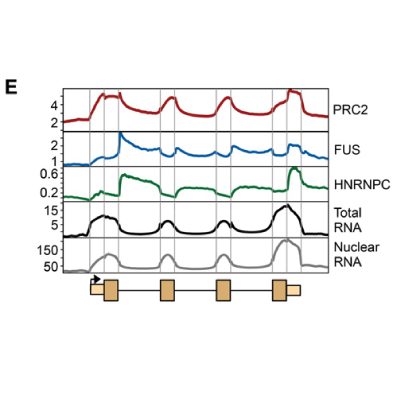
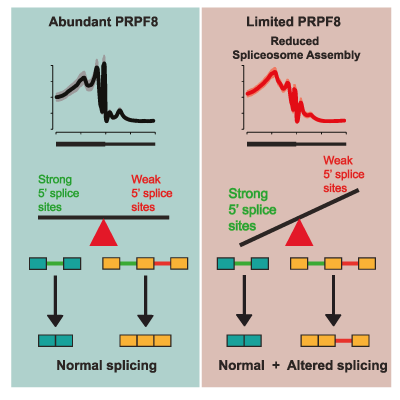
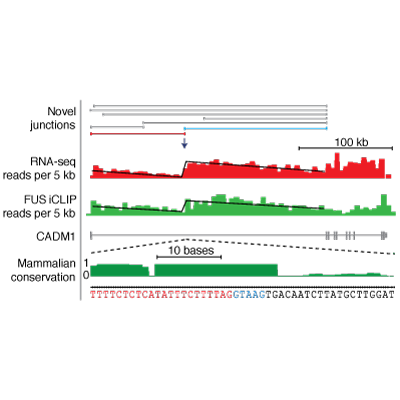
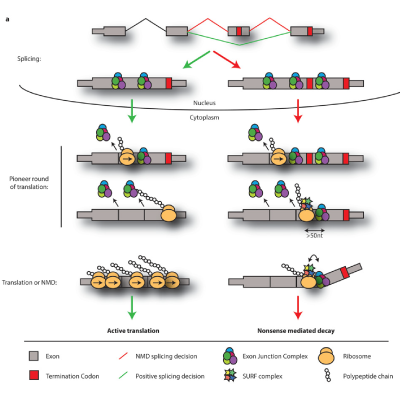
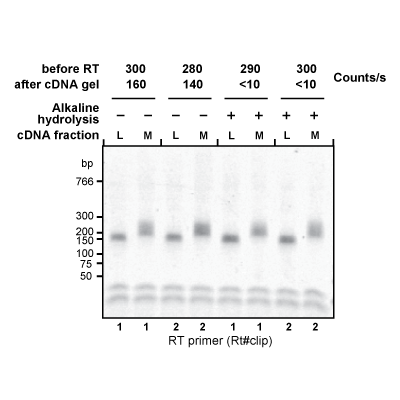
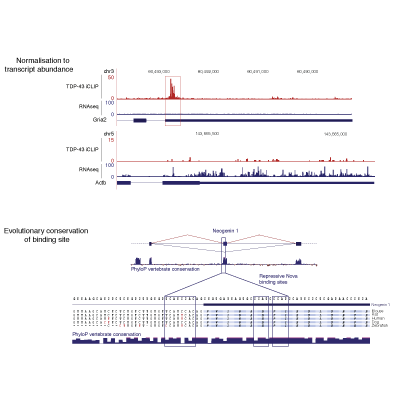
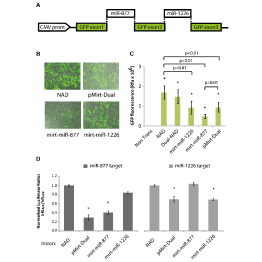
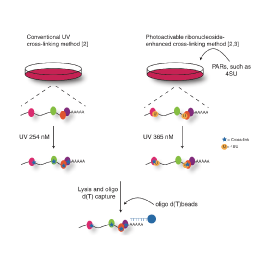
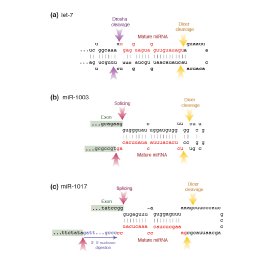
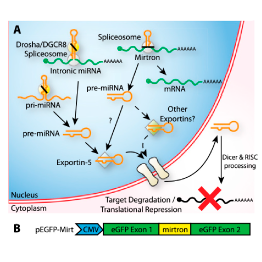
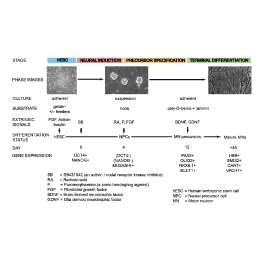
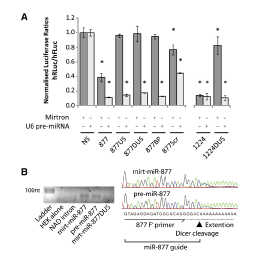
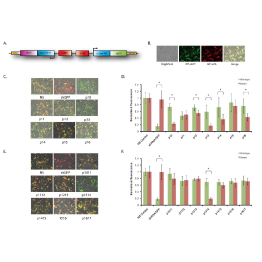
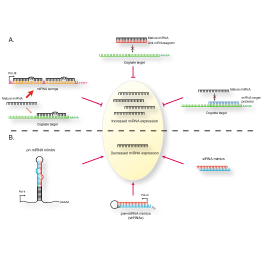
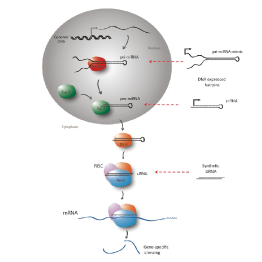
Master Regulators
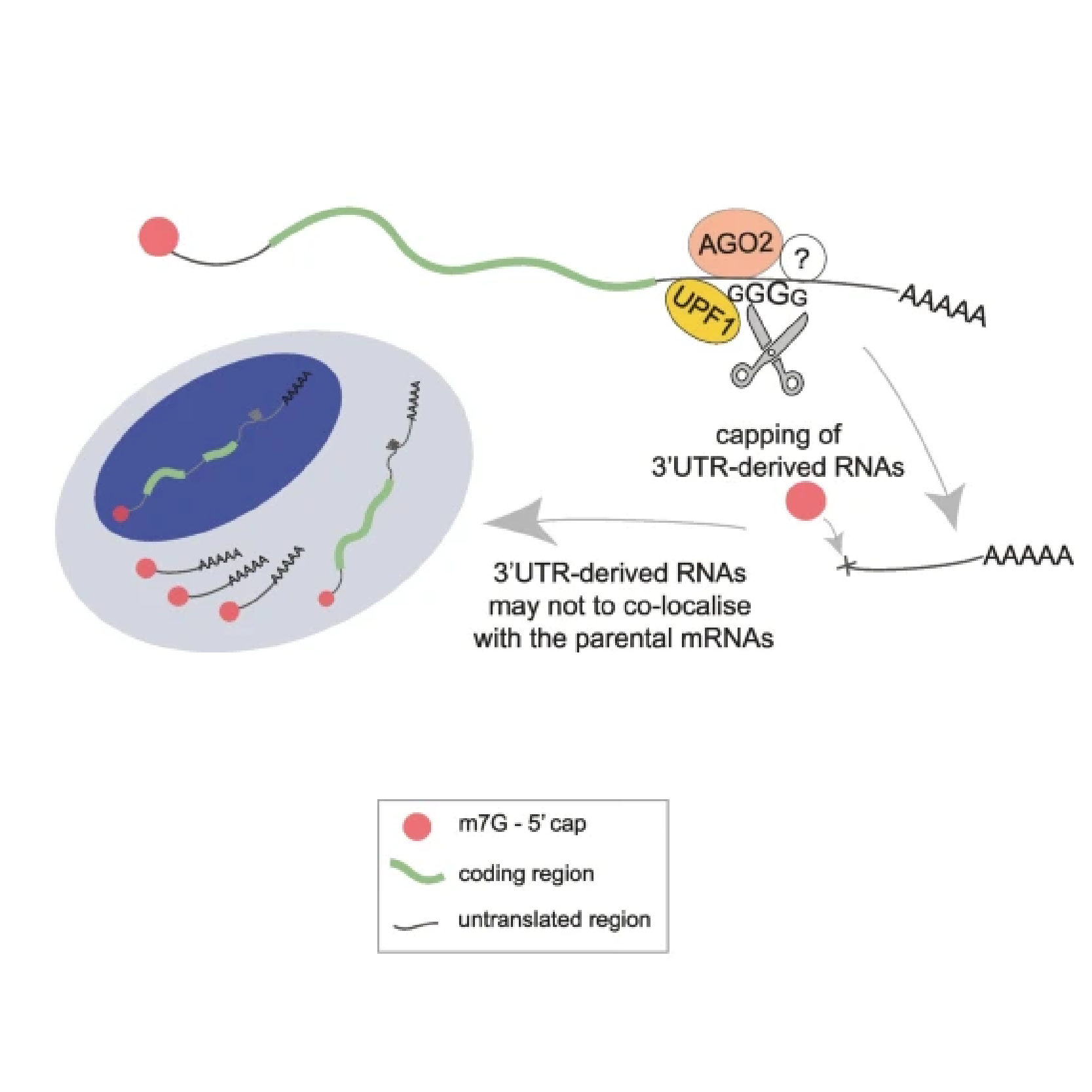
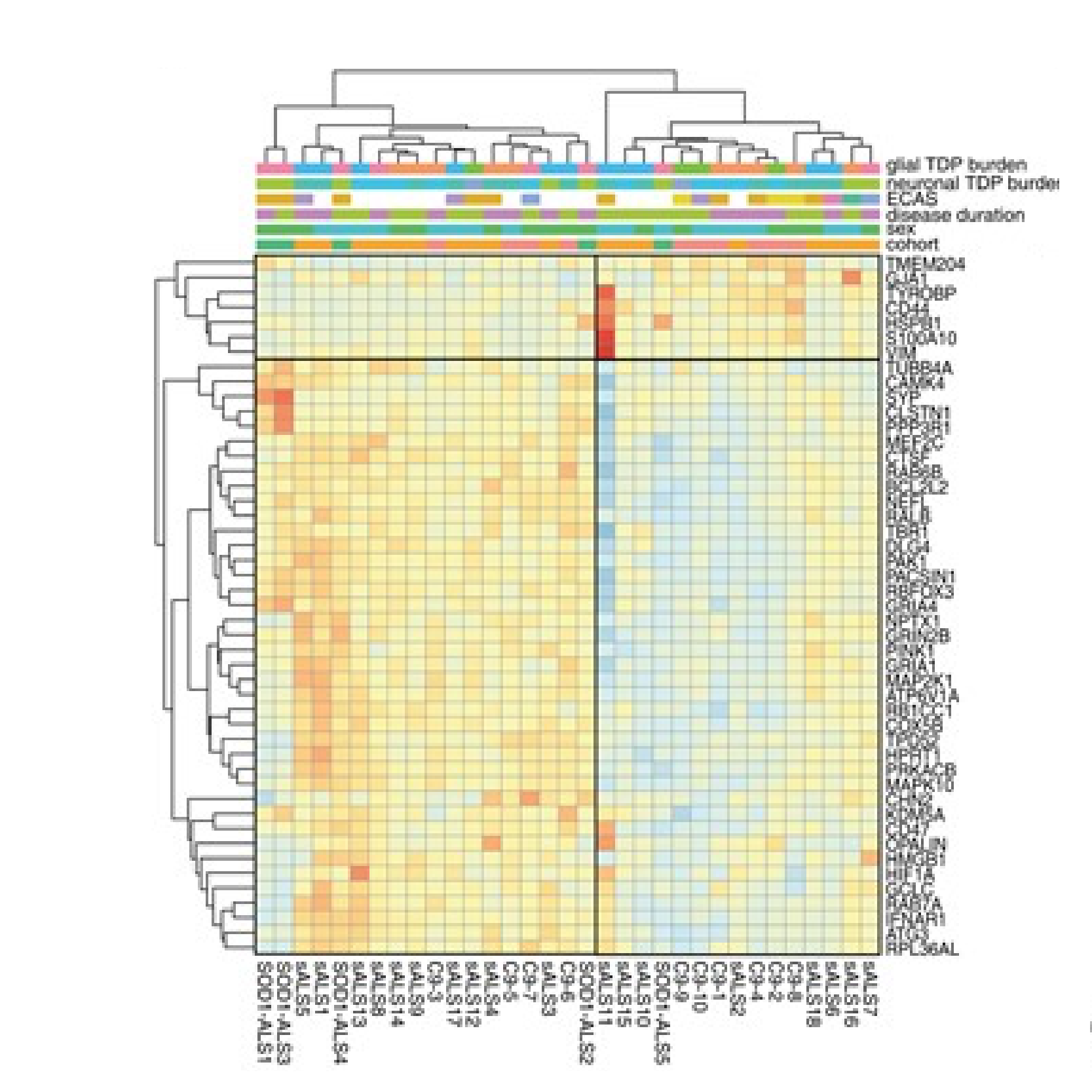
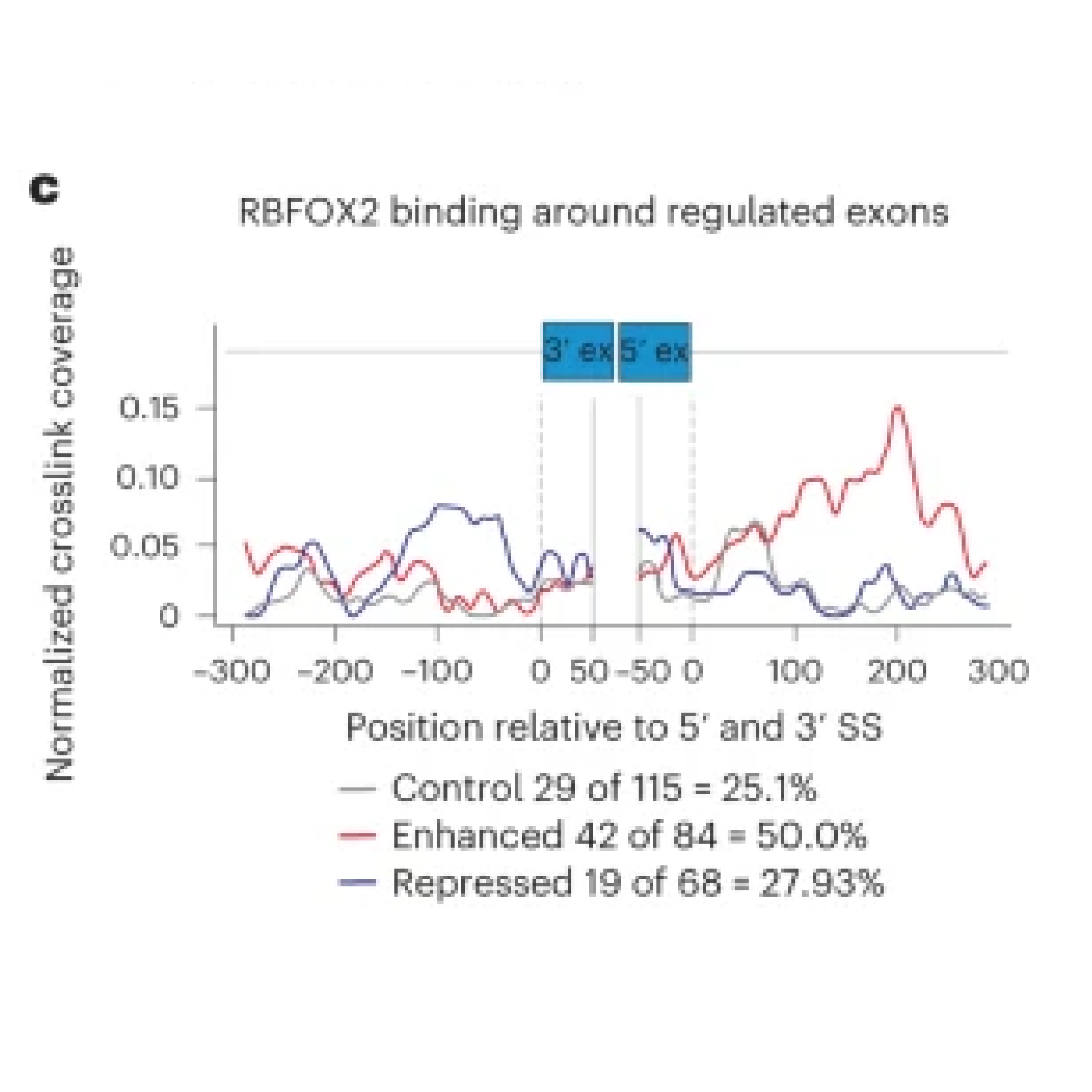
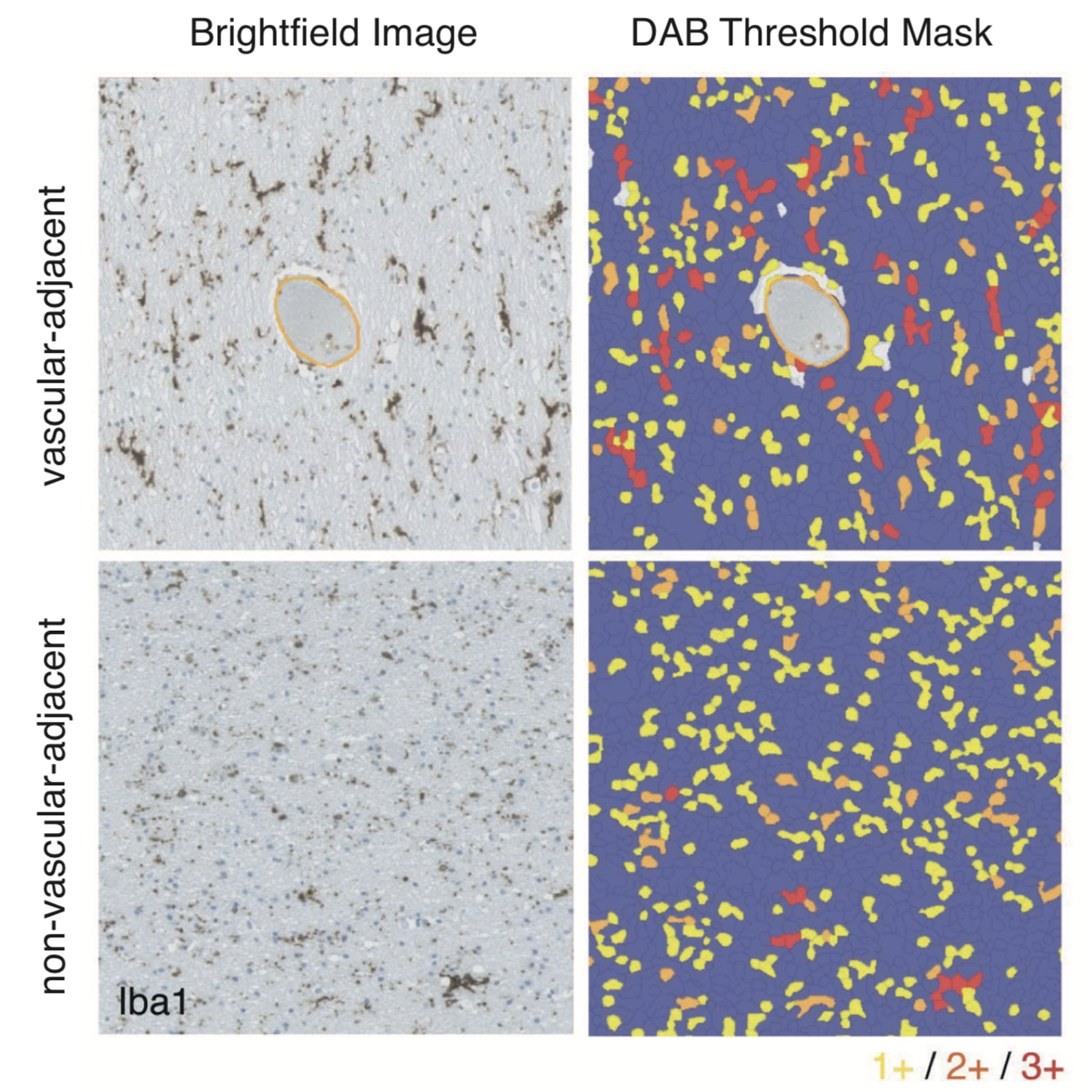
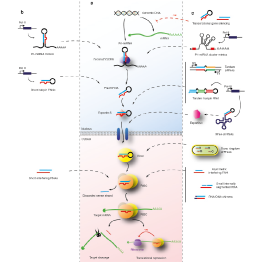
A main focus of the group is to use a combination of functional genomics and computational approaches to research master regulators in both post-mortem tissue and human cell models of neurological disease. Specifically, we define and explore target networks of both transcription factors and RNA-binding proteins. This is with the aim of identifying target networks that are perturbed in disease and which contribute to neurological disease phenotypes. In parallel, we are using computational models to understand how genomic variation impacts on regulator-target networks. Ultimately we plan to use our network models to predict the interactions that might be corrected to allow phenotypic recovery.